ADVERTISEMENT
ORFs, Splicing & Coding
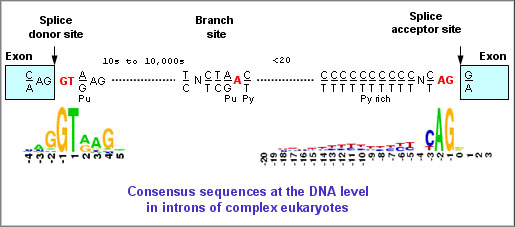
ORFs, Start Sites & Coding Regions
The ORF Finder (Open Reading Frame Finder) is a graphical analysis tool which finds all open reading frames of a selectable minimum size in a user's sequence or in a sequence already in the database.
This server provides access to the program Genscan for predicting the locations and exon-intron structures of genes in genomic sequences from a variety of organisms. This server can accept sequences up to 1 million base pairs (1 Mbp) in length.
The NetStart server produces neural network predictions of translation start in vertebrate and Arabidopsis thaliana nucleotide sequences.
Projector is a program for the comparative, homology based prediction of protein coding genes in mouse and human DNA. Projector takes the known genes of one DNA sequence and predicts the corresponding genes in an evolutionarily related DNA sequence.
Online ORF analysis giving detailed report.
HMM-based gene structure prediction (multiple genes, both chains)
Protea is a software devoted to protein-coding sequences identification. The input is a set of DNA sequences that need not to be aligned. The method takes advantage of the specific substitution pattern of coding sequences together with the consistency of reading frames.
Gene identification in novel eukaryotic genomes by self-training algorithm
Splicing, Alternative Splicing & Introns
Ecgene is novel gene prediction program that combined genome-based EST clustering and transcript assembly procedures in a coherent fashion andprovides a genome annotaion for alternative splicing.
Computes scores at Donor and Acceptor site; identifies Branch Point (BP) and Polypyrimidine Tract (PPT) sites in an intronic sequence.
This server provides access to the program GenomeScan for predicting the locations and exon-intron structures of genes in genomic sequences from a variety of organisms.
TassDB stores extensive data about alternative splice events at GYNGYN donors and NAGNAG acceptors. Currently, 114,554 tandem splice sites of eight species are contained in the database, 5,209 of which have EST/mRNA evidence for alternative splicing.
The AStalavista web server extracts and displays alternative splicing (AS) events from a given genomic annotation of exon-intron gene coordinates.
This web interface provides a tool to predict the effects of sequence changes that alter mRNA splicing in human diseases. We designed the system to evaluate changes in splice site strength based on information theory-based models of donor and acceptor splice sites.
ASSP predicts putative alternative exon isoform, cryptic, and constitutive splice sites of internal (coding) exons.
A fast, flexible system for detecting splice sites in the genomic DNA of various eukaryotes.
The NetGene2 server is a service producing neural network predictions of splice sites in human, C. elegans and A. thaliana DNA.
Several examples of intronic and exonic cis-elements that are important for correct splice-site identification and are distinct from the classical splicing signals have been described. These elements can act both by stimulating (enhancers) or by repressing (silencers) splicing, and they seem to be especially relevant for regulating alternative splicing. Exonic splicing enhancers (ESEs), in particular, appear to be very prevalent, and may be present in most, if not all exons, including constitutive ones
The NetPlantGene server is a service producing neural network predictions of splice sites in Arabidopsis thaliana DNA.
SpliceCenter is a suite of user-friendly tools that evaluate the impact of gene splicing variation on specific molecular biology techniques.
Group II introns are a novel class of RNAs best known for their self-splicing reaction. Under certain in vitro conditions, the introns can excise themselves from precursor mRNAs and ligate together their flanking exons, without the aid of protein.
Intron & Exon Databases
List of Gene structure, introns and exons, splice sites Databases at Nucleic Acid Research.
Hollywood exon annotation database at MIT, a website for querying a relational database of constitutive and alternative human exons, by using biological and descriptive features.
Human-transcriptome DataBase of Alternative Splicing
DBTSS is a database of transcriptional start sites, based on our unique collection of precise, experimentally-determined 5'-end sequences of full-length cDNAs.
HS3D (Homo Sapiens Splice Sites Dataset) is a data set of Homo Sapiens Exon, Intron and Splice regions extracted from GenBank
his site contains information about the spliceosomal introns of the yeast Saccharomyces cerevisiae.
The Web Bench
The Web Bench
is the essential companion to the biologist, bringing informational resources and a collection of tools & calculators to facilitate work at the bench and analysis of biological data.
Check out the full online bench here
Check out the full online bench here
Sequence Analysis with GenBeans
 Try GenBeans: Best free software for DNA sequence editing!
Try GenBeans: Best free software for DNA sequence editing!
FEEDBACK
Your comments & your suggestionsare appreciated. Please, notify us for resources and tools that you would like to see on this bench!





