DNA & Nucleotides Properties
- Nucleic Acid Molecular Weight
- Oligonucleotide Concentration
- DNA Spectrophotometric Conversions
- Molar Conversions for common plasmids
- DNA Coding capacity
- DNA End Concentration
- Nucleotide Properties
- Nucleotide Conversion Formula
- Base Pair Structure
Nucleic Acid MW ssDNA (e.g., Oligonucleotides) MW = (An x 313.2) + (Tn x 304.2) + (Cn x 289.2) + (Gn x 329.2) + 79.0 (5’-P if any) An, Tn, Cn, and Gn number of each nucleotide. Average MW of a base = 330 (with sodium) dsDNA Average MW of a DNA base pair = 660 (610 without sodium) Average dsDNA MW = 660 x N (N – number of base pairs)
Oligonucleotide Concentration C (µM or pmol/µl) = A260 / (0.01 x N) C (ng/ml) = (A260 x MW) / (0.01 x N) MW - molecular weight or relative molecular mass (Mr) A260 - absorbance at 260nm N - number of bases
DNA Spectrophotometric Conversions ds DNA: 1.0 A260 unit = 50 µg/ml = 0.15 mM (in nucleotides) ss DNA: 1.0 A260 unit = 33 µg/ml = 0.10 mM (in nucleotides) 1 mM dsDNA (in nucleotides) = 6.7 A260 units 1 mM ssDNA (in nucleotides) = 10.0 A260 units
Molar Conversions for common plasmids
1 µg of 1000 bp DNA = 1.52 pmol
= 9.1 X 1011 molecules
1 µg of pUC18/19 DNA (2686 bp) = 0.57 pmol
= 3.4 X 10(11) molecules
1 µg of pBR322 DNA (4361 bp) = 0.35 pmol
= 2.1 X 10(11) molecules
1 µg of SV40 DNA (5243 bp) = 0.29 pmol
1 µg of PhiX174 DNA (5386 bp) = 0.28 pmol
1 µg of M13mp18/19 DNA (7249 bp) = 0.21 pmol
= 1.3 X 10(11) molecules
1 µg of lambda phage DNA (48502 bp) = 0.03 pmol
= 1.8 X 10(10) molecules
1 pmol of 1000 bp DNA = 0.66 µg
1 pmol of pUC18/19 DNA (2686 bp) = 1.77 µg
1 pmol of pBR322 DNA (4361 bp) = 2.88 µg
1 pmol of SV40 DNA (5243 bp) = 3.46 µg
1 pmol of PhiX174 DNA (5386 bp) = 3.54 µg
1 pmol of M13mp18/19 DNA (7250 bp) = 4.78 µg
1 pmol of lambda phage DNA (48502 bp) = 32.01 µg
DNA Coding capacity 1.0 kb DNA = 333 amino acids 10 kDa protein @ 270 bp DNA 37 kDa protein @ 1.0 kb DNA 50 kDa protein @ 1.35 kb DNA
DNA End Concentration ends (pmol) = DNA (pmol) x (number of cuts x 2) Add one cut if the DNA was already linear before cleavage 1 µg of 1000 bp DNA = 3.04 pmol ends 1 µg of linearized pUC18/19 DNA = 1.14 pmol ends 1 µg of linearized pBR322 DNA = 0.7 pmol ends 1 µg of linearized SV40 DNA = 0.58 pmol ends 1 µg of linearized PhiX174 DNA = 0.56 pmol ends 1 µg of linearized M13mp18/19 DNA = 0.42 pmol ends 1 µg of linearized lambda phage DNA = 0.06 pmol ends
Nucleotide Properties Compound MW λmax (nm) e (M-1 x cm-1) Ribonucleotide Triphosphates (Avg. MW = 499.5) ATP 507.2 259 15,400 CTP 483.2 271 9,000 GTP 523.2 253 13,700 UTP 484.2 262 10,000 Deoxyribonucleotide triphosphates (Avg. MW = 487.0) dATP 491.2 259 15,200 dCTP 467.2 271 9,300 dGTP 507.2 253 13,700 dTTP 482.2 267 9,600 lambda max determined at pH 7.0 e = molar absorption coefficient = absorbance at lambda max for 1 M solution at pH 7.0.
Nucleotide Conversion Formula C = A / e x 10(3), where C - mM concentration of compounds A - absorbance at λmax (OD) for 1 cm cuvette e - molar absorption coefficient (M-1 x cm-1)
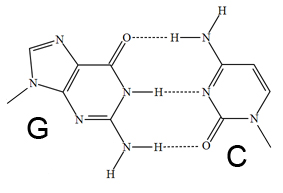
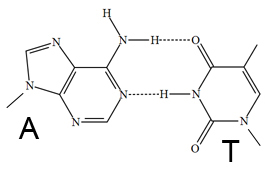
The Web Bench
The Web Bench
is the essential companion to the biologist, bringing informational resources and a collection of tools & calculators to facilitate work at the bench and analysis of biological data.
Check out the full online bench here
Check out the full online bench here
Sequence Analysis with GenBeans
 Try GenBeans: Best free software for DNA sequence editing!
Try GenBeans: Best free software for DNA sequence editing!
FEEDBACK
Your comments & your suggestionsare appreciated. Please, notify us for resources and tools that you would like to see on this bench!





